Getting Started
FabIO is a Python module for reading and handling data from two-dimensional X-ray detectors.
FabIO is a Python module written for easy and transparent reading
of raw two-dimensional data from various X-ray detectors. The
module provides a function for reading any image and returning a
FabioImage object which contains both metadata (header information)
and the raw data. All FabioImage object offer additional methods to
extract information about the image and to open other detector
images from the same data series.
Introduction
One obstacle when writing software to analyse data collected from a two-dimensional detector is to read the raw data into the program, not least because the data can be stored in many different formats depending on the instrument used. To overcome this problem we decided to develop a general module, FabIO (FABle I/O), to handle reading and writing of two-dimensional data. The code-base was initiated by merging parts of our fabian imageviewer and ImageD11 peak-search programs and has been developed since 2007 as part of the TotalCryst program suite for analysis of 3DXRD microscopy data. During integration into a range of scientific programs like the FABLE graphical interface, EDNA and the fast azimuthal integration library, pyFAI; FabIO has gained several features like handling multi-frame image formats as well as writing many of the file formats.
FabIO Python module
Python is a scripting language that is very popular among scientists and which also allows well structured applications and libraries to be developed.
Philosophy
The intention behind this development was to create a Python module which would enable easy reading of 2D data images, from any detector without having to worry about the file format. Therefore FabIO just needs a file name to open a file and it determines the file format automatically and deals with gzip and bzip2 compression transparently. Opening a file returns an object which stores the image in memory as a 2D NumPy array and the metadata, called header, in a Python dictionary. Beside the data and header attributes, some methods are provided for reading the previous or next image in a series of images as well as jumping to a specific file number. For the user, these auxiliary methods are intended to be independent of the image format (as far as is reasonably possible).
FabIO is written in an object-oriented style (with classes) but aims at being used in a scripting environment: special care has been taken to ensure the library remains easy to use. Therefore no knowledge of object-oriented programming is required to get full benefits of the library. As the development is done in a collaborative and decentralized way; a comprehensive test suite has been added to reduce the number of regressions when new features are added or old problems are repaired. The software is very modular and allows new classes to be added for handling other data formats easily. FabIO and its source-code are freely available to everyone on-line, licensed under the liberal MIT License. FabIO is also available directly from popular Linux distributions like Debian and Ubuntu.
Implementation
The main language used in the development of FabIO is Python; however, some image formats are compressed and require compression algorithms for reading and writing data. When such algorithms could not be implemented efficiently using Python or NumPy native modules were developed, in i.e. standard C code callable from Python (sometimes generated using Cython). This code has to be compiled for each computer architecture and offers excellent performance. FabIO is only dependent on the NumPy module and has extra features if two other optional Python modules are available. For reading XML files (that are used in EDNA) the lxml module is required and the Python Image Library, PIL is needed for producing a PIL image for displaying the image in graphical user interfaces and several image-processing operations that are not re-implemented in FabIO. A variety of useful image processing is also available in the scipy.ndimage module and in scikits-image.
Images can also be displayed in a convenient interactive manner using matplotlib and an IPython shell , which is mainly used for developing data analysis algorithms. Reading and writing procedure of the various TIFF formats is based on the TiffIO code from PyMCA.
In the Python shell, the fabio module must be imported prior to reading an image in one of the supported file formats (see Table Supported formats, hereafter). The fabio.open function creates an instance of the Python class fabioimage, from the name of a file. This instance, named img hereafter, stores the image data in img.data as a 2D NumPy array. Often the image file contains more information than just the intensities of the pixels, e.g. information about how the image is stored and the instrument parameters at the time of the image acquisition, these metadata are usually stored in the file header. Header information, are available in img.header as a Python dictionary where keys are strings and values are usually strings or numeric values.
Information in the header about the binary part of the image (compression, endianness, shape) are interpreted however, other metadata are exposed as they are recorded in the file. FabIO allows the user to modify and, where possible, to save this information (the table Supported formats summarizes writable formats). Automatic translation between file-formats, even if desirable, is sometimes impossible because not all format have the capability to be extended with additional metadata. Nevertheless FabIO is capable of converting one image data-format into another by taking care of the numerical specifics: for example float arrays are converted to integer arrays if the output format only accepts integers.
FabIO methods
One strength of the implementation in an object oriented language is the possibility to combine functions (or methods) together with data appropriate for specific formats.
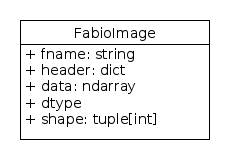
In addition to the header information and
image data,
every FabioImage instance (returned
by fabio.open()) has methods inherited from FabioImage which
provide information about the image minimum, maximum and mean
values.
import fabio
image = fabio.open('image.tif')
print(image.header) # print the header
print(image.data.mean()) # print mean intensity of the data
image.close()
FabIO old-fashion file series
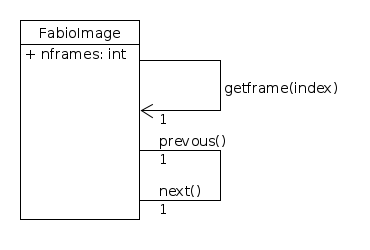
Multi-frames for certain file formats are handled using file series.
A set of methodes, specific for certain formats provide access to the data
through a series of files. These methods are next(),
previous(), and getframe().
The behaviour of such methods varies depending on the image format: for single-frame
format (like mar345), next() will return the image in next
file; for multi-frame format (like GE), next() will return
the next frame within the same file. For formats which are possibly
multi-framed like EDF, the behaviour depends on the actual number
of frames per file (accessible via the nframes attribute).
import fabio
im100 = fabio.open('Quartz_0100.tif') # Open image file
print(im0.data[1024,1024]) # Check a pixel value
im101 = im100.next() # Open next image
im270 = im100.getframe(270) # Jump to file number 270: Quartz_0270.tif
This conveniant way to iterate through many files have limitation. It is not working in case of many frames per files, the read access is difficult to optimize, and it is difficult to custom the list of the files.
FabIO file series
This design introduces a real FabioFrame as composition
of all FabioImage.
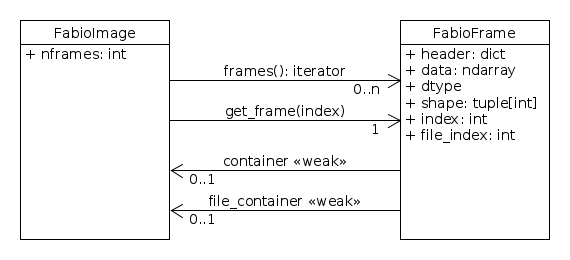
For single frames images, FabioImage still provides
access to the data of to the first (and only one) frame. But the method get_frame()
(with an underscore, not getframe()) provides access to any frames contained in the file.
A file containing a single data, also contains a single frame object. Both provide the same data
(there is 2 ways to access to this data).
To iterate other many files a FileSeries can be used.
This object is a FabioImage which a set of file as a single container of frame.
The hi-level function open_series() is provided to hide the complexity.
This function (or this class) allows different ways to custom the file iteration. Plus optional informnation to describe the way frames as stored in files in order to optimize the random access.
Methodes provided allow to reach frames
using a sequencial access (frames())
or using a random access (get_frame()).
# Random access
import fabio
# The first filename of consecutive filenames while foobar_xxxx.edf exists
filename = "foobar_0000.edf"
with fabio.open_series(first_filename=filename) as series:
frame1 = series.get_frame(1)
frame100 = series.get_frame(100)
frame19 = series.get_frame(19)
Usually, in case of a random access only accessed data have to be decoded, but the file have to be fully read to index the frames (depending of the codec, then the file format).
In case of huge EDF file series a sequencial access to the frames speed up by 2 the reading time.
# Sequencial access
import fabio
# The first filename of consecutive filenames while foobar_xxxx.edf exists
filename = "foobar_0000.edf"
with fabio.open_series(first_filename=filename) as series:
for frame in series.frames():
frame.data
frame.header
frame.index # frame index inside the file series
frame.file_index # frame index inside the edf file
frame.file_container.filename # name of the source file
Examples
Normalising the intensity to a value in the header
img = fabio.open('exampleimage0001.edf')
print(img.header)
{'ByteOrder': 'LowByteFirst',
'DATE (scan begin)': 'Mon Jun 28 21:22:16 2010',
'ESRFCurrent': '198.099',
...
}
# Normalise to beam current and save data
srcur = float(img.header['ESRFCurrent'])
img.data *= 200.0/srcur
img.write('normed_0001.edf')
Interactive viewing with matplotlib
from matplotlib import pyplot # Load matplotlib
pyplot.imshow(img.data) # Display as an image
pyplot.show() # Show GUI window
Converting a TIFF to an EDF
import fabio
image = fabio.open("my.tiff")
image.convert("edf").save("my.edf")
Future and perspectives
The Hierarchical Data Format version 5 (hdf5) is a data format which is increasingly popular for storage of X-ray and neutron data. To name a few facilities the synchrotron Soleil and the neutron sources ISIS, SNS and SINQ already use HDF extensively through the NeXus format. For now, mainly processed or curated data are stored in this format but new detectors (Eiger from Dectris) are natively saving data in HDF5. FabIO will rely on H5Py, which already provides a good HDF5 binding for Python, as an external dependency, to be able to read and write such HDF5 files. This starts to be available in version 0.4.0.
Conclusion
FabIO gives an easy way to read and write 2D images when using the Python computer language. It was originally developed for X-ray diffraction data but now gives an easy way for scientists to access and manipulate their data from a wide range of 2D X-ray detectors. We welcome contributions to further improve the code and hope to add more file formats in the future.
Acknowledgements
We acknowledge Andy Götz and Kenneth Evans for extensive testing when including the FabIO reader in the Fable image viewer (Götz et al., 2007). We also thank V. Armando Solé for assistance with his TiffIO reader and Carsten Gundlach for deployment of FabIO at the beamlines i711 and i811, MAX IV, and providing bug reports. We finally acknowledge our colleagues who have reported bugs and helped to improve FabIO. Financial support was granted by the EU 6th Framework NEST/ADVENTURE project TotalCryst (Poulsen et al., 2006).
Citation
Knudsen, E. B., Sørensen, H. O., Wright, J. P., Goret, G. & Kieffer, J. (2013). J. Appl. Cryst. 46, 537-539.
List of file formats that FabIO can read and write
In alphabetical order. The listed filename extensions are typical examples. FabIO tries to deduce the actual format from the file itself and only uses extensions as a fallback if that fails.
Python Module |
Detector / Format |
Extension |
Read |
Multi-image |
Write |
|---|---|---|---|---|---|
ADSC |
ADSC Quantum |
.img |
Yes |
No |
Yes |
Bruker86 |
Bruker formats |
.sfrm |
Yes |
No |
Yes |
Bruker100 |
Bruker formats |
.sfrm |
Yes |
No |
Yes |
CBF |
CIF binary files |
.cbf |
Yes |
No |
Yes |
DM3 |
Gatan Digital Micrograph |
.dm3 |
Yes |
No |
No |
EDF |
ESRF data format |
.edf |
Yes |
Yes |
Yes |
EDNA-XML |
Used by EDNA |
.xml |
Yes |
No |
No |
Eiger |
Dectris format |
.h5 |
Yes |
Yes |
Yes |
Fit2D |
Fit2D binary format |
.f2d |
Yes |
No |
No |
Fit2D mask |
Fit2D mask |
.msk |
Yes |
No |
Yes |
Fit2D spreadsheet |
Fit2D Ascii format |
.spr |
Yes |
No |
Yes |
GE |
General Electric |
Yes |
Yes |
No |
|
JPEG |
Joint Photographic Experts Group |
.jpg |
Yes |
No |
No |
JPEG2k |
JPEG 2000 |
.jpx |
Yes |
No |
No |
Hdf5 |
Needs the dataset path |
.h5 |
Yes |
Yes |
No |
HiPiC |
Hamamatsu CCD |
.tif |
Yes |
No |
No |
kcd |
Nonius KappaCCD |
.kccd |
Yes |
No |
No |
marccd |
MarCCD/Mar165 |
.mccd |
Yes |
No |
No |
mar345 |
Mar345 image plate |
.mar3450 |
Yes |
No |
Yes |
mpa |
Multi-wire detector |
.mpa |
yes |
No |
No |
mrc |
Medical Research Council |
.map |
Yes |
Yes |
No |
numpy |
numpy 2D array |
.npy |
Yes |
No |
Yes |
OXD |
Oxford Diffraction |
.img |
Yes |
No |
Yes |
Pixi |
pixi |
Yes |
No |
No |
|
pilatus |
Dectris Pilatus Tiff |
.tif |
Yes |
No |
Yes |
PNM |
Portable aNy Map |
.pnm |
Yes |
No |
Yes |
Raxis |
Rigaku Saxs format |
.img |
Yes |
No |
No |
spe |
Princeton instrumentation |
.spe |
Yes |
Yes |
No |
TIFF |
Tagged Image File Format |
.tif |
Yes |
Yes |
Yes |
Adding new file formats
We hope it will be relatively easy to add new file formats to FabIO in the future. Please refere at the fabio/templateimage.py file in the source which describes how to add a new format.