Cologne Database for Molecular Spectroscopy (CDMS) Queries (astroquery.linelists.cdms)¶
Getting Started¶
The CDMS module provides a query interface for the Search and Conversion Form
of the Cologne Database for Molecular Spectroscopy. The module outputs the results that
would arise from the browser form using similar search criteria as the ones
found in the form, and presents the output as a Table. The
module is similar in spirit and content to the JPLSpec module.
Examples¶
Querying the catalog¶
The default option to return the query payload is set to False. In the
following examples we have explicitly set it to False and True to show the what
each setting yields:
>>> from astroquery.linelists.cdms import CDMS
>>> import astropy.units as u
>>> response = CDMS.query_lines(min_frequency=100 * u.GHz,
max_frequency=1000 * u.GHz,
min_strength=-500,
molecule="028503 CO",
get_query_payload=False)
>>> print(response)
FREQ ERR LGINT DR ELO GUP TAG QNFMT Ju Ku vu Jl Kl vl F name
MHz MHz MHz nm2 1 / cm
----------- ------ ------- --- -------- --- ------ ----- --- --- --- --- --- --- --- -------
115271.2018 0.0005 -7.1425 2 0.0 3 -28503 101 1 -- -- -- -- -- 0 CO, v=0
230538.0 0.0005 -6.1605 2 3.845 5 -28503 101 2 -- -- -- -- -- 1 CO, v=0
345795.9899 0.0005 -5.6026 2 11.535 7 -28503 101 3 -- -- -- -- -- 2 CO, v=0
461040.7682 0.0005 -5.2128 2 23.0695 9 -28503 101 4 -- -- -- -- -- 3 CO, v=0
576267.9305 0.0005 -4.9132 2 38.4481 11 -28503 101 5 -- -- -- -- -- 4 CO, v=0
691473.0763 0.0005 -4.6701 2 57.6704 13 -28503 101 6 -- -- -- -- -- 5 CO, v=0
806651.806 0.005 -4.4657 2 80.7354 15 -28503 101 7 -- -- -- -- -- 6 CO, v=0
921799.7 0.005 -4.2895 2 107.6424 17 -28503 101 8 -- -- -- -- -- 7 CO, v=0
The following example, with get_query_payload = True, returns the payload:
>>> response = CDMS.query_lines(min_frequency=100 * u.GHz,
max_frequency=1000 * u.GHz,
min_strength=-500,
molecule="028503 CO",
get_query_payload=True)
>>> print(response)
[('MinNu', 100.0), ('MaxNu', 1000.0), ('UnitNu', 'GHz'), ('StrLim', -500),
('temp', 0), ('logscale', 'yes'), ('mol_sort_query', 'tag'), ('sort',
'frequency'), ('output', 'text'), ('but_action', 'Submit'), ('Molecules',
'028503 CO')]
The units of the columns of the query can be displayed by calling
response.info:
>>> response = CDMS.query_lines(min_frequency=100 * u.GHz,
max_frequency=1000 * u.GHz,
min_strength=-500,
molecule="028503 CO",
get_query_payload=False)
>>> print(response.info)
<Table length=8>
name dtype unit class n_bad
----- ------- ------- ------------ -----
FREQ float64 MHz Column 0
ERR float64 MHz Column 0
LGINT float64 MHz nm2 Column 0
DR int64 Column 0
ELO float64 1 / cm Column 0
GUP int64 Column 0
TAG int64 Column 0
QNFMT int64 Column 0
Ju int64 Column 0
Ku int64 MaskedColumn 8
vu int64 MaskedColumn 8
Jl int64 MaskedColumn 8
Kl int64 MaskedColumn 8
vl int64 MaskedColumn 8
F int64 Column 0
name str7 Column 0
These come in handy for converting to other units easily, an example using a simplified version of the data above is shown below:
>>> print (response)
FREQ ERR ELO
MHz MHz 1 / cm
----------- ------- -------
115271.2018 0.0005 0.0
345795.9899 0.0005 11.535
461040.7682 0.0005 23.0695
>>> response['FREQ'].quantity
<Quantity [115271.2018,345795.9899,461040.7682] MHz>
>>> response['FREQ'].to('GHz')
<Quantity [115.2712018,345.7959899,461.0407682] GHz>
The parameters and response keys are described in detail under the Reference/API section.
Looking Up More Information from the catdir.cat file¶
If you have found a molecule you are interested in, the TAG field in the results provides enough information to access specific molecule information such as the partition functions at different temperatures. Keep in mind that a negative TAG value signifies that the line frequency has been measured in the laboratory but not in space
>>> import matplotlib.pyplot as plt
>>> from astroquery.linelists.cdms import CDMS
>>> result = CDMS.get_species_table()
>>> mol = result[result['tag'] == 28503] #do not include signs of TAG for this
>>> print(mol)
tag molecule #lines lg(Q(1000)) lg(Q(500)) lg(Q(300)) lg(Q(225)) lg(Q(150)) lg(Q(75)) lg(Q(37.5)) lg(Q(18.75)) lg(Q(9.375)) lg(Q(5.000)) lg(Q(2.725))
----- -------- ------ ----------- ---------- ---------- ---------- ---------- --------- ----------- ------------ ------------ ------------ ------------
28503 CO, v=0 95 2.5595 2.2584 2.0369 1.9123 1.7370 1.4386 1.1429 0.8526 0.5733 0.3389 0.1478
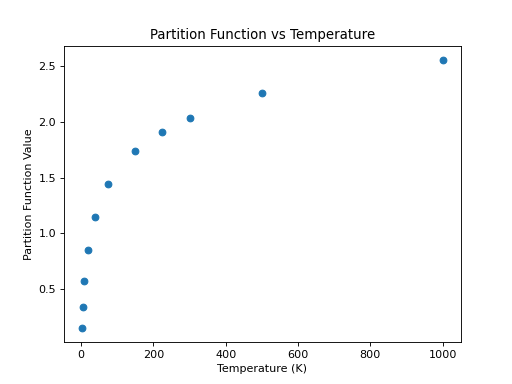
One of the advantages of using CDMS is the availability in the catalog of the partition function at different temperatures for the molecules (just like for JPL). As a continuation of the example above, an example that accesses and plots the partition function against the temperatures found in the metadata is shown below:
>>> keys = [k for k in mol.keys() if 'lg' in k]
>>> temp = np.array([float(k.split('(')[-1].split(')')[0]) for k in keys])
>>> part = list(mol[keys][0])
>>> plt.scatter(temp,part)
>>> plt.xlabel('Temperature (K)')
>>> plt.ylabel('Partition Function Value')
>>> plt.title('Partition Function vs Temperature')
(Source code, png, hires.png, pdf)
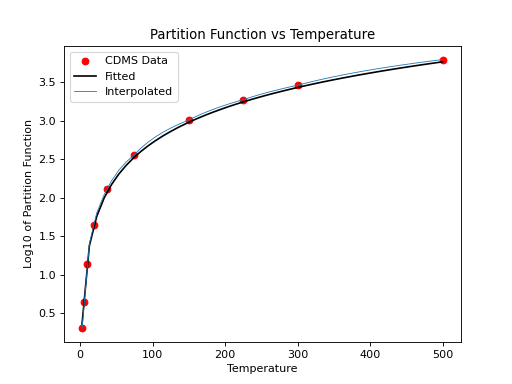
For non-linear molecules like H2CO, curve fitting methods can be used to
calculate production rates at different temperatures with the proportionality:
a*T**(3./2.). Calling the process above for the H2CO molecule (instead of
for the CO molecule) we can continue to determine the partition function at
other temperatures using curve fitting models:
>>> result = CDMS.get_species_table()
>>> mol = result[result['TAG'] == 30501] #do not include signs of TAG for this
>>> print(mol)
>>> from scipy.optimize import curve_fit
>>> def f(T, a):
return np.log10(a*T**(1.5))
>>> keys = [k for k in mol.keys() if 'lg' in k]
>>> def tryfloat(x):
try:
return float(x)
except:
return np.nan
>>> temp = np.array([float(k.split('(')[-1].split(')')[0]) for k in keys])
>>> part = np.array([tryfloat(x) for x in mol[keys][0]])
>>> param, cov = curve_fit(f, temp[np.isfinite(part)], part[np.isfinite(part)])
>>> print(param)
# array([0.51865074])
>>> x = np.linspace(2.7,500)
>>> y = f(x,param[0])
>>> plt.scatter(temp,part,c='r')
>>> plt.plot(x,y,'k')
>>> plt.title('Partition Function vs Temperature')
>>> plt.xlabel('Temperature')
>>> plt.ylabel('Log10 of Partition Function')
(Source code, png, hires.png, pdf)
We can then compare linear interpolation to the fitted interpolation above:
>>> interp_Q = np.interp(x, temp, 10**part)
>>> plt.plot(x, (10**y-interp_Q)/10**y)
>>> plt.xlabel("Temperature")
>>> plt.ylabel("Fractional difference between linear and fitted")
(Source code, png, hires.png, pdf)
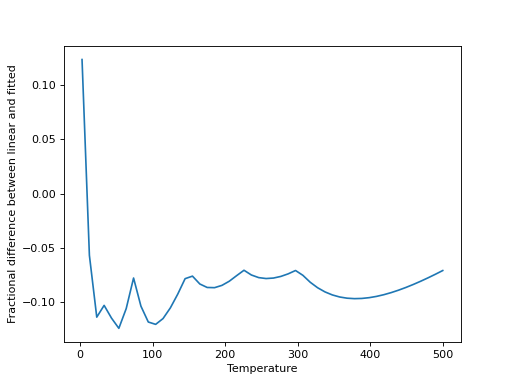
Linear interpolation is a good approximation, in this case, for any moderately high temperatures, but is increasingly poor at lower temperatures. It can be valuable to check this for any given molecule.
Querying the Catalog with Regexes and Relative names¶
The regular expression parsing is analogous to that in the JPLSpec module.
Reference/API¶
astroquery.linelists.cdms Package¶
CDMS catalog¶
Cologne Database for Molecular Spectroscopy query tool
Classes¶
|
Configuration parameters for |