Calculating Delta-values¶
In this tutorial we compare the equation-of-state (EOS) calculated for 7 FCC
metals using values from EMT, WIEN2k and
experiment. Each EOS is described by three parameters:
volume per atom
bulk-modulus
pressure derivative of bulk-modulus
Differences between two EOS’es can be measured by a single \(\Delta\) value defined as:
where \(E_n(V)\) is the energy per atom as a function of volume.
The \(\Delta\) value can be calculated using the
ase.utils.deltacodesdft.delta() function:
- ase.utils.deltacodesdft.delta(v1: float, B1: float, Bp1: float, v2: float, B2: float, Bp2: float, symmetric=True) float[source]¶
Calculate Delta-value between two equation of states.
- Parameters
v1 (float) – Volume per atom.
v2 (float) – Volume per atom.
B1 (float) – Bulk-modulus (in eV/Ang^3).
B2 (float) – Bulk-modulus (in eV/Ang^3).
Bp1 (float) – Pressure derivative of bulk-modulus.
Bp2 (float) – Pressure derivative of bulk-modulus.
symmetric (bool) – Default is to calculate a symmetric delta.
- Returns
delta – Delta value in eV/atom.
- Return type
See also
Collection of ground-state elemental crystals: DeltaCodesDFT
Equation-of-state module:
ase.eos
We get the WIEN2k and experimental numbers from the DeltaCodesDFT ASE-collection and we calculate the EMT EOS using this script:
from ase.calculators.emt import EMT
from ase.collections import dcdft
from ase.io import Trajectory
for symbol in ['Al', 'Ni', 'Cu', 'Pd', 'Ag', 'Pt', 'Au']:
traj = Trajectory('{}.traj'.format(symbol), 'w')
for s in range(94, 108, 2):
atoms = dcdft[symbol]
atoms.set_cell(atoms.cell * (s / 100)**(1 / 3), scale_atoms=True)
atoms.calc = EMT()
atoms.get_potential_energy()
traj.write(atoms)
And fit to a Birch-Murnaghan EOS:
import json
from pathlib import Path
from typing import Tuple
from ase.eos import EquationOfState as EOS
from ase.io import read
def fit(symbol: str) -> Tuple[float, float, float, float]:
V = []
E = []
for atoms in read('{}.traj@:'.format(symbol)):
V.append(atoms.get_volume() / len(atoms))
E.append(atoms.get_potential_energy() / len(atoms))
eos = EOS(V, E, 'birchmurnaghan')
eos.fit(warn=False)
e0, B, Bp, v0 = eos.eos_parameters
return e0, v0, B, Bp
data = {} # Dict[str, Dict[str, float]]
for path in Path().glob('*.traj'):
symbol = path.stem
e0, v0, B, Bp = fit(symbol)
data[symbol] = {'emt_energy': e0,
'emt_volume': v0,
'emt_B': B,
'emt_Bp': Bp}
Path('fit.json').write_text(json.dumps(data))
Result for Pt:
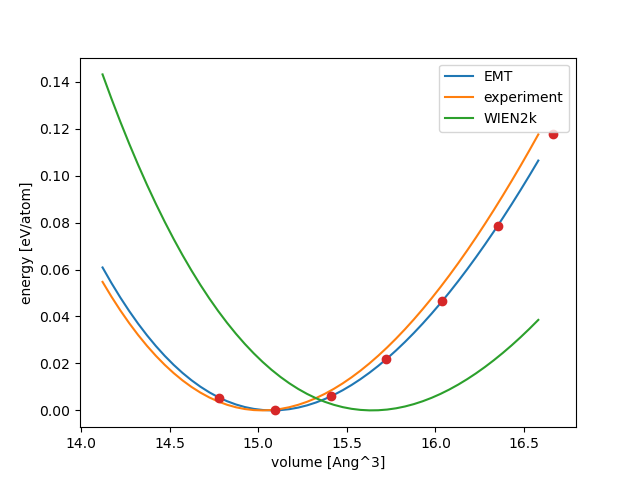
Volumes in Ang^3:
# symbol |
emt |
exp |
wien2k |
Au |
16.68 |
16.82 |
17.97 |
Pt |
15.08 |
15.02 |
15.64 |
Ag |
16.77 |
16.85 |
17.85 |
Pd |
14.59 |
14.56 |
15.31 |
Cu |
11.57 |
11.65 |
11.95 |
Ni |
10.60 |
10.81 |
10.89 |
Al |
15.93 |
16.27 |
16.48 |
Bulk moduli in GPa:
# symbol |
emt |
exp |
wien2k |
Au |
174.12 |
182.01 |
139.11 |
Pt |
278.67 |
285.51 |
248.71 |
Ag |
100.06 |
105.71 |
90.15 |
Pd |
180.43 |
187.19 |
168.63 |
Cu |
134.41 |
144.28 |
141.33 |
Ni |
176.23 |
192.46 |
200.37 |
Al |
39.70 |
77.14 |
78.08 |
Pressure derivative of bulk-moduli:
# symbol |
emt |
exp |
wien2k |
Au |
5.46 |
6.40 |
5.76 |
Pt |
5.31 |
5.18 |
5.46 |
Ag |
4.75 |
4.72 |
5.42 |
Pd |
5.17 |
5.00 |
5.56 |
Cu |
4.21 |
4.88 |
4.86 |
Ni |
3.76 |
4.00 |
5.00 |
Al |
2.72 |
4.45 |
4.57 |
Now, we can calculate \(\Delta\) between EMT and WIEN2k for Pt:
>>> from ase.utils.deltacodesdft import delta
>>> from ase.units import kJ
>>> delta(15.08, 278.67 * 1e-24 * kJ, 5.31,
... 15.64, 248.71 * 1e-24 * kJ, 5.46)
0.03205389052984122
Here are all the values (in meV/atom) calculated with the script below:
# symbol |
emt-exp |
emt-wien2k |
exp-wien2k |
Au |
5.9 |
43.7 |
39.4 |
Pt |
3.5 |
32.2 |
35.9 |
Ag |
1.9 |
22.4 |
21.3 |
Pd |
1.0 |
27.6 |
29.0 |
Cu |
2.7 |
11.9 |
9.5 |
Ni |
8.6 |
12.5 |
3.7 |
Al |
5.9 |
8.6 |
3.6 |
import json
from pathlib import Path
import numpy as np
import matplotlib.pyplot as plt
from ase.collections import dcdft
from ase.eos import birchmurnaghan
from ase.io import read
from ase.units import kJ
from ase.utils.deltacodesdft import delta
# Read EMT data:
data = json.loads(Path('fit.json').read_text())
# Insert values from experiment and WIEN2k:
for symbol in data:
dcdft_dct = dcdft.data[symbol]
dcdft_dct['exp_B'] *= 1e-24 * kJ
dcdft_dct['wien2k_B'] *= 1e-24 * kJ
data[symbol].update(dcdft_dct)
for name in ['volume', 'B', 'Bp']:
with open(name + '.csv', 'w') as f:
print('# symbol, emt, exp, wien2k', file=f)
for symbol, dct in data.items():
values = [dct[code + '_' + name]
for code in ['emt', 'exp', 'wien2k']]
if name == 'B':
values = [val * 1e24 / kJ for val in values]
print('{},'.format(symbol),
', '.join('{:.2f}'.format(value) for value in values),
file=f)
with open('delta.csv', 'w') as f:
print('# symbol, emt-exp, emt-wien2k, exp-wien2k', file=f)
for symbol, dct in data.items():
# Get v0, B, Bp:
emt, exp, wien2k = [(dct[code + '_volume'],
dct[code + '_B'],
dct[code + '_Bp'])
for code in ['emt', 'exp', 'wien2k']]
print('{},'.format(symbol),
'{:.1f}, {:.1f}, {:.1f}'.format(delta(*emt, *exp) * 1000,
delta(*emt, *wien2k) * 1000,
delta(*exp, *wien2k) * 1000),
file=f)
if symbol == 'Pt':
va = min(emt[0], exp[0], wien2k[0])
vb = max(emt[0], exp[0], wien2k[0])
v = np.linspace(0.94 * va, 1.06 * vb)
for (v0, B, Bp), code in [(emt, 'EMT'),
(exp, 'experiment'),
(wien2k, 'WIEN2k')]:
plt.plot(v, birchmurnaghan(v, 0.0, B, Bp, v0), label=code)
e0 = dct['emt_energy']
V = []
E = []
for atoms in read('Pt.traj@:'):
V.append(atoms.get_volume() / len(atoms))
E.append(atoms.get_potential_energy() / len(atoms) - e0)
plt.plot(V, E, 'o')
plt.legend()
plt.xlabel('volume [Ang^3]')
plt.ylabel('energy [eV/atom]')
plt.savefig('Pt.png')