
Image processing in GRASS GIS
Table of contents
- Image processing in general
- Imagery import
- Semantic label information
- Image processing operations
- Geocoding of imagery data
- Visualizing (true) color composites
- Calculation of vegetation indices
- Calibration of thermal channel
- Image classification
- Image fusion
- Radiometric corrections
- Time series processing
- Evapotranspiration modeling
- Energy balance
- See also
Image processing in general
Digital numbers and physical values (reflection/radiance-at-sensor):Satellite imagery is commonly stored in Digital Numbers (DN) for minimizing the storage volume, i.e. the originally sampled analog physical value (color, temperature, etc) is stored a discrete representation in 8-16 bits. For example, Landsat data are stored in 8bit values (i.e., ranging from 0 to 255); other satellite data may be stored in 10 or 16 bits. Having data stored in DN, it implies that these data are not yet the observed ground reality. Such data are called "at-satellite", for example the amount of energy sensed by the sensor of the satellite platform is encoded in 8 or more bits. This energy is called radiance-at-sensor. To obtain physical values from DNs, satellite image providers use a linear transform equation (y = a * x + b) to encode the radiance-at-sensor in 8 to 16 bits. DNs can be turned back into physical values by applying the reverse formula (x = (y - b) / a).
The GRASS GIS module i.landsat.toar easily transforms Landsat DN to radiance-at-sensor (top of atmosphere, TOA). The equivalent module for ASTER data is i.aster.toar. For other satellites, r.mapcalc can be employed.
Reflection/radiance-at-sensor and surface reflectance
When radiance-at-sensor has been obtained, still the atmosphere influences the signal as recorded at the sensor. This atmospheric interaction with the sun energy reflected back into space by ground/vegetation/soil needs to be corrected. The need of removing atmospheric artifacts stems from the fact that the atmosphericic conditions are changing over time. Hence, to gain comparability between Earth surface images taken at different times, atmospheric need to be removed converting at-sensor values which are top of atmosphere to surface reflectance values.
In GRASS GIS, there are two ways to apply atmospheric correction for satellite imagery. A simple, less accurate way for Landsat is with i.landsat.toar, using the DOS correction method. The more accurate way is using i.atcorr (which supports many satellite sensors). The atmospherically corrected sensor data represent surface reflectance, which ranges theoretically from 0% to 100%. Note that this level of data correction is the proper level of correction to calculate vegetation indices.
In GRASS GIS, image data are identical to raster data. However, a couple of commands are explicitly dedicated to image processing. The geographic boundaries of the raster/imagery file are described by the north, south, east, and west fields. These values describe the lines which bound the map at its edges. These lines do NOT pass through the center of the grid cells at the edge of the map, but along the edge of the map itself.
As a general rule in GRASS:
- Raster/imagery output maps have their bounds and resolution equal to those of the current region.
- Raster/imagery input maps are automatically cropped/padded and rescaled (using nearest-neighbor resampling) to match the current region.
Imagery import
The module r.in.gdal offers a common interface for many different raster and satellite image formats. Additionally, it also offers options such as on-the-fly location creation or extension of the default region to match the extent of the imported raster map. For special cases, other import modules are available. Always the full map is imported. Imagery data can be group (e.g. channel-wise) with i.group.For importing scanned maps, the user will need to create a x,y-location, scan the map in the desired resolution and save it into an appropriate raster format (e.g. tiff, jpeg, png, pbm) and then use r.in.gdal to import it. Based on reference points the scanned map can be rectified to obtain geocoded data.
Semantic label information
Semantic labels are a description which can be stored as metadata. To print available semantic labels relevant for multispectral satellite data, use i.band.library. r.semantic.label allows assigning of these satellite imagery band references as defined in i.band.library. Semantic labels are also used in signature files of imagery classification tools. Therefore, signature files of one imagery or raster group can be used to classify a different group with identical semantic labels.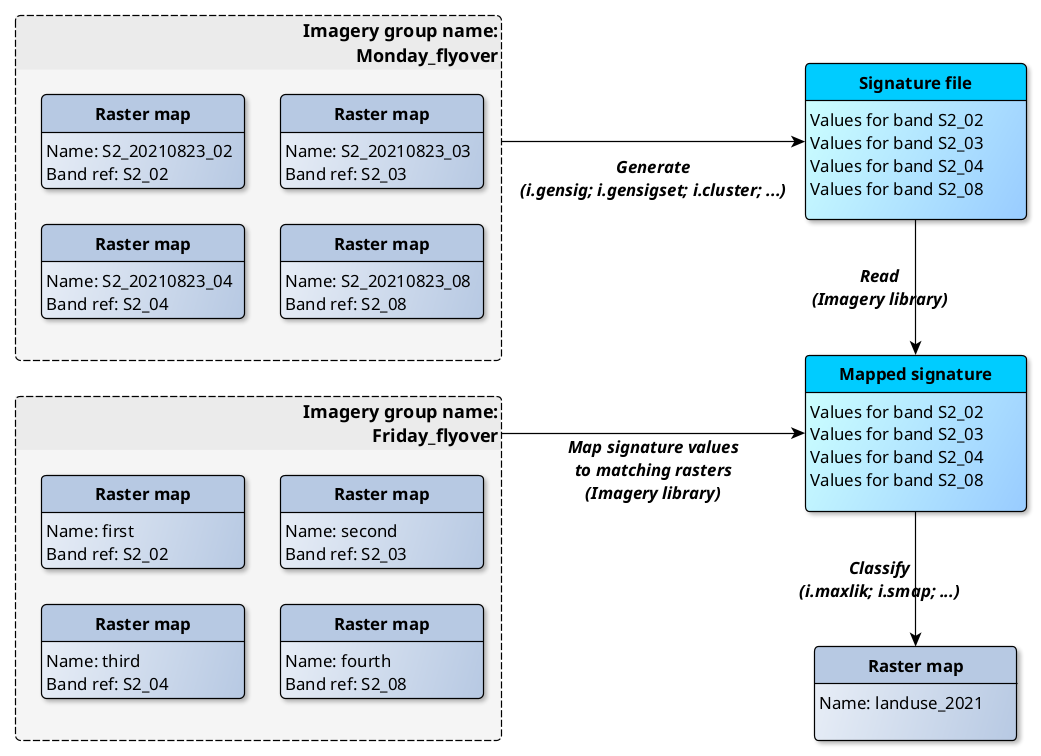
New enhanced classification workflow involving semantic labels.
Image processing operations
GRASS raster/imagery map processing is always performed in the current region settings (see g.region), i.e. the current region extent and current raster resolution is used. If the resolution differs from that of the input raster map(s), on-the-fly resampling is performed (nearest neighbor resampling). If this is not desired, the input map(s) has/have to be resampled beforehand with one of the dedicated modules.Geocoding of imagery data
GRASS is able to geocode raster and image data of various types:- unreferenced scanned maps by defining four corner points (i.group, i.target, g.gui.gcp, i.rectify)
- unreferenced satellite data from optical and Radar sensors by defining a certain number of ground control points (i.group, i.target, g.gui.gcp, i.rectify)
- interactive graphical Ground Control Point (GCP) manager
- orthophoto generation based on DEM: i.ortho.photo
- digital handheld camera geocoding: modified procedure for i.ortho.photo
Visualizing (true) color composites
To quickly combine the first three channels to a near natural color image, the GRASS command d.rgb can be used or the graphical GIS manager (wxGUI). It assigns each channel to a color which is then mixed while displayed. With a bit more work of tuning the grey scales of the channels, nearly perfect colors can be achieved. Channel histograms can be shown with d.histogram.Calculation of vegetation indices
An example for indices derived from multispectral data is the NDVI (normalized difference vegetation index). To study the vegetation status with NDVI, the Red and the Near Infrared channels (NIR) are taken as used as input for simple map algebra in the GRASS command r.mapcalc (ndvi = 1.0 * (nir - red)/(nir + red)). With r.colors an optimized "ndvi" color table can be assigned afterward. Also other vegetation indices can be generated likewise.Calibration of thermal channel
The encoded digital numbers of a thermal infrared channel can be transformed to degree Celsius (or other temperature units) which represent the temperature of the observed land surface. This requires a few algebraic steps with r.mapcalc which are outlined in the literature to apply gain and bias values from the image metadata.Image classification
Single and multispectral data can be classified to user defined land use/land cover classes. In case of a single channel, segmentation will be used. GRASS supports the following methods:- Radiometric classification:
- Unsupervised classification (i.cluster, i.maxlik) using the Maximum Likelihood classification method
- Supervised classification (i.gensig or g.gui.iclass, i.maxlik) using the Maximum Likelihood classification method
- Combined radiometric/geometric (segmentation based) classification:
- Supervised classification (i.gensigset, i.smap)
- Object-oriented classification:
- Unsupervised classification (segmentation based: i.segment)
Note - signatures generated for one scene are suitable for classification of other scenes as long as they consist of same raster bands (semantic labels match). This comes handy when classifying multiple scenes from a single sensor taken in different areas or different times.
Image fusion
In case of using multispectral data, improvements of the resolution can be gained by merging the panchromatic channel with color channels. GRASS provides the HIS (i.rgb.his, i.his.rgb) and the Brovey and PCA transform (i.pansharpen) methods.Radiometric corrections
Atmospheric effects can be removed with i.atcorr. Correction for topographic/terrain effects is offered in i.topo.corr. Clouds in LANDSAT data can be identified and removed with i.landsat.acca. Calibrated digital numbers of LANDSAT and ASTER imagery may be converted to top-of-atmosphere radiance or reflectance and temperature (i.aster.toar, i.landsat.toar).Time series processing
GRASS also offers support for time series processing (r.series). Statistics can be derived from a set of coregistered input maps such as multitemporal satellite data. The common univariate statistics and also linear regression can be calculated.Evapotranspiration modeling
In GRASS, several types of evapotranspiration (ET) modeling methods are available:- Reference ET: Hargreaves (i.evapo.mh), Penman-Monteith (i.evapo.pm);
- Potential ET: Priestley-Taylor (i.evapo.pt);
- Actual ET: i.evapo.time.
Energy balance
Emissivity can be calculated with i.emissivity. Several modules support the calculation of the energy balance:- Actual evapotranspiration for diurnal period (i.eb.eta);
- Evaporative fraction and root zone soil moisture (i.eb.evapfr);
- Sensible heat flux iteration (i.eb.hsebal01);
- Net radiation approximation (i.eb.netrad);
- Soil heat flux approximation (i.eb.soilheatflux).
See also
- GRASS GIS Wiki page: Image processing
- The GRASS 4 Image Processing manual
- Introduction into raster data processing
- Introduction into 3D raster data (voxel) processing
- Introduction into vector data processing
- Introduction into temporal data processing
- Database management
- Projections and spatial transformations
SOURCE CODE
Available at: Image processing in GRASS GIS source code (history)
Accessed: Saturday Jan 21 17:40:37 2023
Main index | Imagery index | Topics index | Keywords index | Graphical index | Full index
© 2003-2023 GRASS Development Team, GRASS GIS 8.2.1 Reference Manual