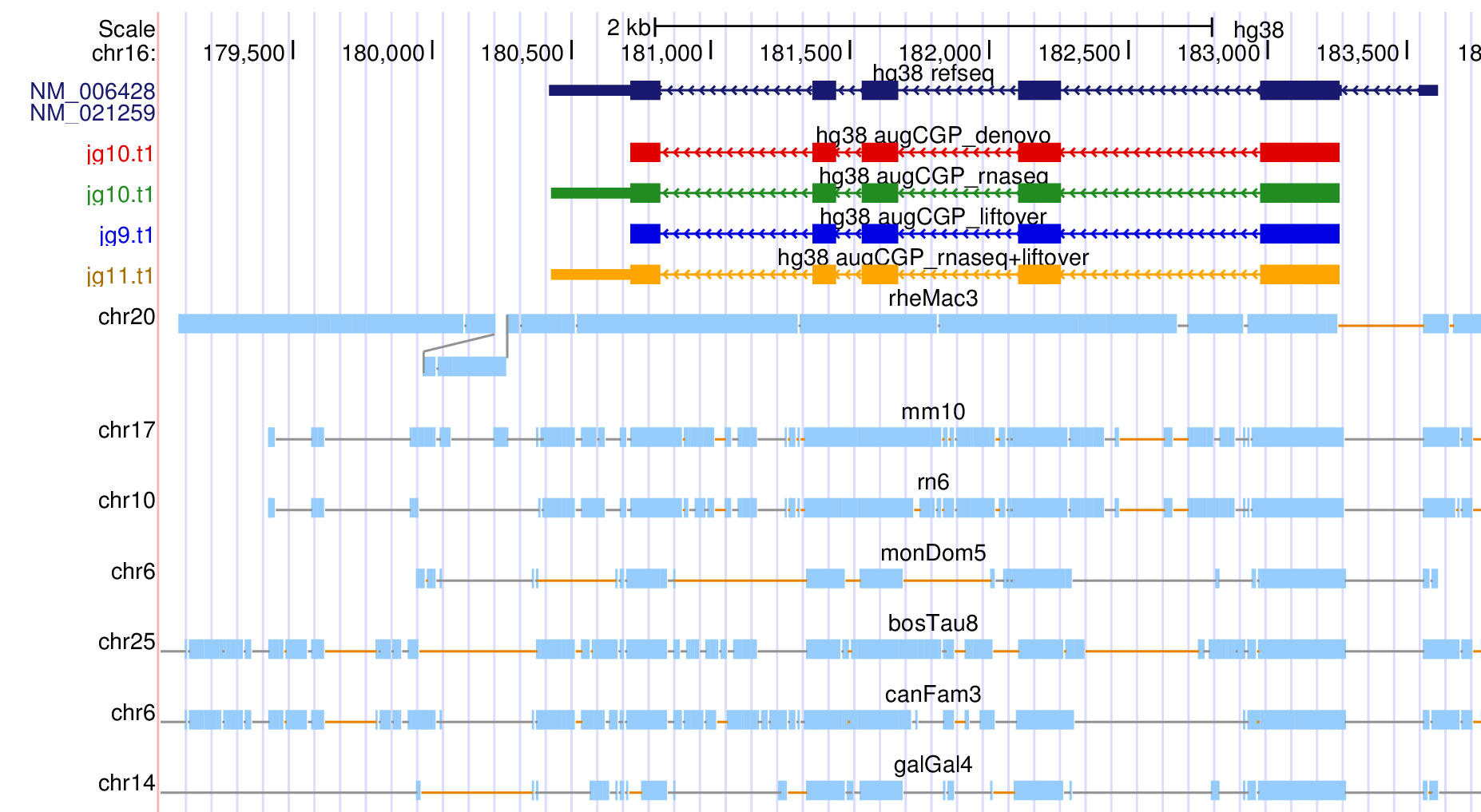
In this tutorial we practice the most common applications of AUGUSTUS-CGP: de novo gene finding (i.e. only the raw genomes are used and no extrinsic evidence), integration of RNA-Seq evidence and lifting over annotations from one (or more) species to the other species in the clade. AUGUSTUS-CGP has three mandatory inputs: the set of genomes, each in Multi-FASTA format, an alignment of the genomes and a phylogenetic tree in NEWICK format. You don't have a whole-genome alignment for your clade? No problem - the tutorial covers how a whole-genome alignment can be created with progressiveCactus. The output alignment of progressiveCactus is in HAL format, which can be displayed as Assembly Hubs in the Genome Browser. In one of the exercise, we will set up such an Assembly Hub and show how gene tracks can be uploaded for visualization. If the phylogeny of the species is not known, we recommend using a star-like tree with uniform branch lengths.