Markov Chain¶
This example builds a Markov Chain
Code¶
import opengm
import numpy
chainLength=5
numLabels=100
numberOfStates=numpy.ones(chainLength,dtype=opengm.label_type)*numLabels
gm=opengm.gm(numberOfStates,operator='adder')
#add some random unaries
for vi in range(chainLength):
unaryFuction=numpy.random.random(numLabels)
gm.addFactor(gm.addFunction(unaryFuction),[vi])
#add one 2.order function
f=opengm.differenceFunction(shape=[numLabels]*2,weight=0.1)
print type(f),f
fid=gm.addFunction(f)
#add factors on a chain
for vi in range(chainLength-1):
gm.addFactor(fid,[vi,vi+1])
inf = opengm.inference.BeliefPropagation(gm,parameter=opengm.InfParam(steps=40,convergenceBound=0 ,damping=0.9))
inf.infer(inf.verboseVisitor())
print inf.arg()
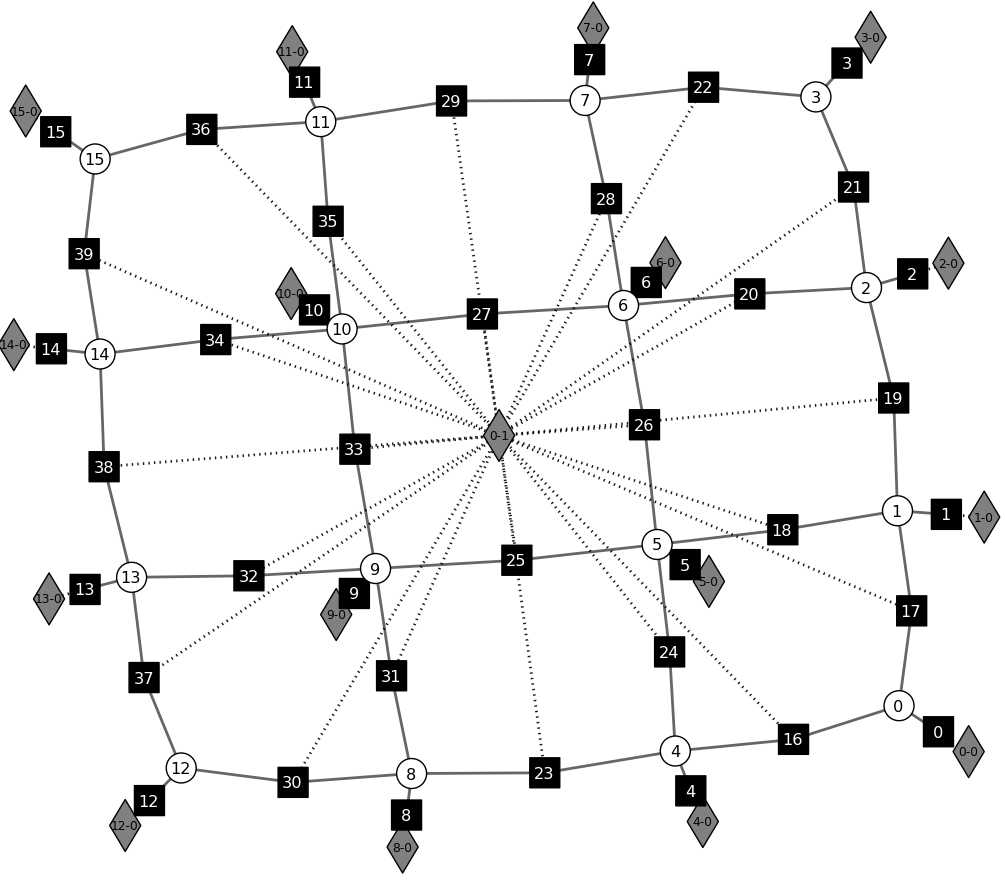
Extended Factor Graph¶
An auto-generated [1] graph [2] for this graphical model.
See also
opengm.visualizeGm() opengm.__init__.grid2d2Order() opengm.grid2d2Order()
Potts Model¶
This example builds a Potts model on a two dimensional grid
Code¶
import numpy
import opengm
img=numpy.random.rand(4,4)
dimx=img.shape[0]
dimy=img.shape[1]
numVar=dimx*dimy
numLabels=2
beta=0.3
numberOfStates=numpy.ones(numVar,dtype=opengm.index_type)*numLabels
gm=opengm.graphicalModel(numberOfStates,operator='adder')
#Adding unary function and factors
for y in range(dimy):
for x in range(dimx):
f=numpy.ones(2,dtype=numpy.float32)
f[0]=img[x,y]
f[1]=1.0-img[x,y]
fid=gm.addFunction(f)
gm.addFactor(fid,(x*dimy+y,))
#Adding binary function and factors"
vis=numpy.ones(5,dtype=opengm.index_type)
#add one binary function (potts fuction)
f=numpy.ones(pow(numLabels,2),dtype=numpy.float32).reshape(numLabels,numLabels)*beta
for l in range(numLabels):
f[l,l]=0
fid=gm.addFunction(f)
#add binary factors
for y in range(dimy):
for x in range(dimx):
if(x+1<dimx):
#vi as tuple (list and numpy array can also be used as vi's)
gm.addFactor(fid,numpy.array([x*dimy+y,(x+1)*dimy+y],dtype=opengm.index_type))
if(y+1<dimy):
#vi as list (tuple and numpy array can also be used as vi's)
gm.addFactor(fid,[x*dimy+y,x*dimy+(y+1)])
opengm.hdf5.saveGraphicalModel(gm,'/tmp/model.h5','gm')
icm=opengm.inference.Icm(gm)
icm.infer()
argmin=icm.arg()
res=argmin.reshape(dimx,dimy)
Interpixel-Boundary Segmentation’-Model¶
This example builds a ‘interpixel-boundary segmentation’ model on a two dimensional grid
Note
TODO: show input and output image for different betas !
Code¶
import opengm
import numpy
import sys
class TopologicalCoordinateToIndex:
def __init__(self ,geometricGridSize) :
self.gridSize=geometricGridSize
def convert(self, tx,ty) :
return tx / 2 + (ty / 2)*(self.gridSize[0]) + ((ty + ty % 2) / 2)*(self.gridSize[0] - 1)
def printSolution(data,solution,coordinateHelper):
for x in range (data.shape[0]*2 - 1):
sys.stdout.write("___")
sys.stdout.write("\n")
for y in range (data.shape[1]*2 - 1):
sys.stdout.write("|")
for x in range (data.shape[0]*2 - 1):
if x % 2 == 0 and y % 2 == 0:
sys.stdout.write(" ")
elif x % 2 == 0 and y % 2 == 1 :
if solution[coordinateHelper.convert(x,y)]==1:
sys.stdout.write("___")
else:
sys.stdout.write(" ")
elif x % 2 == 1 and y % 2 == 0 :
if solution[coordinateHelper.convert(x,y)]==1:
sys.stdout.write(" | ")
else:
sys.stdout.write(" ")
elif x % 2 == 1 and y % 2 == 1:
sys.stdout.write(" * ")
sys.stdout.write("|\n")
for x in range (data.shape[1]*2 - 1):
sys.stdout.write("___")
sys.stdout.write("\n")
# model parameter
gridSize=[4,4] # size of grid
beta=0.7 # bias to choose between under- and over-segmentation
high=100 # closedness-enforcing soft-constraint value for forbidden configurations
# size of the topological grid
tGridSize=[2*gridSize[0] -1,2*gridSize[1] -1]
nrOfVariables=gridSize[1]*(gridSize[0]-1)+gridSize[0]*(gridSize[1]-1)
cToVi=TopologicalCoordinateToIndex(gridSize)
# some random data on a grid
data=numpy.random.random(gridSize[0]*gridSize[1]).astype(numpy.float32).reshape(gridSize[0],gridSize[1])
# construct gm
numberOfLabels=numpy.ones(nrOfVariables,dtype=opengm.label_type)*2
gm=opengm.graphicalModel(numberOfLabels)
# 4th closedness-function
fClosedness=numpy.zeros( pow(2,4),dtype=numpy.float32).reshape(2,2,2,2)
for x1 in range(2):
for x2 in range(2):
for x3 in range(2):
for x4 in range(2):
labelsum=x1+x2+x3+x4
if labelsum is not 2 and labelsum is not 0 :
fClosedness[x1,x2,x3,x4]=high
fidClosedness=gm.addFunction(fClosedness)
# for each boundary in the grid, i.e. for each variable
# of the model, add one 1st order functions
# and one 1st order factor
# and for each junction of four inter-pixel edges on the grid,
# one factor is added that connects the corresponding variable
# indices and refers to the closedness-function
for yt in range(tGridSize[1]):
for xt in range(tGridSize[0]):
# unaries
if (xt % 2 + yt % 2) == 1 :
gradient = abs( data[xt / 2, yt / 2]- data[xt/ 2 + xt % 2, yt / 2 + yt % 2])
f=numpy.array([beta*gradient , (1.0-beta)*(1.0-gradient)])
gm.addFactor(gm.addFunction(f),[cToVi.convert(xt,yt)])
# high order factors (4.th order)
if xt % 2 + yt % 2 == 2 :
vi=[cToVi.convert(xt + 1, yt),cToVi.convert(xt - 1, yt),cToVi.convert(xt, yt + 1), cToVi.convert(xt, yt - 1)]
vi=sorted(vi);
gm.addFactor(fidClosedness,vi)
inf=opengm.inference.LazyFlipper(gm,parameter=opengm.InfParam(maxSubgraphSize=4))
inf.inference.infer()
arg=inf.inference.arg()
printSolution(data,arg,cToVi)
Pure Python ICM Solver¶
This example show how to write inference solvers in python, and shows the usage of the movemaker
Code¶
import opengm
import numpy
unaries=numpy.random.rand(100 , 100,2)
potts=opengm.PottsFunction([2,2],0.0,0.2)
gm=opengm.grid2d2Order(unaries=unaries,regularizer=potts)
class IcmPurePython():
def __init__(self,gm):
self.gm=gm
self.numVar=gm.numberOfVariables
self.movemaker=opengm.movemaker(gm)
self.adj=gm.variablesAdjacency()
self.localOpt=numpy.zeros(self.numVar,dtype=numpy.bool)
def infer(self,verbose=False):
changes=True
while(changes):
changes=False
for v in self.gm.variables():
if(self.localOpt[v]==False):
l=self.movemaker.label(v)
nl=self.movemaker.moveOptimallyMin(v)
self.localOpt[v]=True
if(nl!=l):
if(verbose):print self.movemaker.value()
self.localOpt[self.adj[v]]=False
changes=True
def arg(self):
argLabels=numpy.zeros(self.numVar,dtype=opengm.label_type)
for v in self.gm.variables():
argLabels[v]=self.movemaker.label(v)
return argLabels
icm=IcmPurePython(gm)
icm.infer(verbose=False)
arg=icm.arg()
print arg.reshape(100,100)
Iterate over a factors value table with a ‘shapeWalker’¶
This example show how to iterate over all possible labelings of a factor
import opengm
import numpy
# some graphical model
# -with 3 variables with 4 labels.
# -with 2 random 2-order functions
# -connected to 2 factors
gm=opengm.gm([4]*3)
f=numpy.random.rand(4,4).astype(numpy.float32)
gm.addFactor(gm.addFunction(f),[0,1])
f=numpy.random.rand(4,4).astype(numpy.float32)
gm.addFactor(gm.addFunction(f),[1,2])
# iterate over all factors of the graphical model
for factor in gm.factors():
# iterate over all labelings with a "shape walker"
for coord in opengm.shapeWalker(f.shape):
print " f[",coord,"]=",factor[coord]
Pure Python Visitors¶
This example shows how to write a pure python visitor for inference. In this example the current state of the solver is reshaped as a 2d image since the model is a 2d grid.
Code¶
"""
Usage: python_visitor_gui.py
This script shows how one can implement visitors
in pure python and inject them into OpenGM solver.
( not all OpenGM solvers support this kind of
code injection )
"""
import opengm
import numpy
import matplotlib
from matplotlib import pyplot as plt
shape=[100,100]
numLabels=10
unaries=numpy.random.rand(shape[0], shape[1],numLabels)
potts=opengm.PottsFunction([numLabels,numLabels],0.0,0.4)
gm=opengm.grid2d2Order(unaries=unaries,regularizer=potts)
inf=opengm.inference.BeliefPropagation(gm,parameter=opengm.InfParam(damping=0.5))
class PyCallback(object):
def __init__(self,shape,numLabels):
self.shape=shape
self.numLabels=numLabels
self.cmap = matplotlib.colors.ListedColormap ( numpy.random.rand ( self.numLabels,3))
matplotlib.interactive(True)
def begin(self,inference):
print "begin of inference"
def end(self,inference):
print "end of inference"
def visit(self,inference):
gm=inference.gm()
labelVector=inference.arg()
print "energy ",gm.evaluate(labelVector)
labelVector=labelVector.reshape(self.shape)
plt.imshow(labelVector*255.0, cmap=self.cmap,interpolation="nearest")
plt.draw()
callback=PyCallback(shape,numLabels)
visitor=inf.pythonVisitor(callback,visitNth=1)
inf.infer(visitor)
argmin=inf.arg()
MRF - Benchmark¶
Mrf-Benchmark Models implemented in Opengm Python:
Stereo matching problems:¶
Note
This example will be there soon.
Photomontage problems:¶
Note
This example will be there soon.
Binary image segmentation problems:¶
Note
This example will be there soon.
Denoising and Inpainting problems:¶
Code:¶
# FIXMEEEEEEEEEEE
import opengm
import vigra # only to read images
import numpy
#import sys
# to animate the current labeling matplotlib is used
import matplotlib
import matplotlib.pyplot as plt
import matplotlib.cm as cm
from matplotlib import animation
class PyCallback(object):
"""
callback functor which will be passed to an inference
visitor.
In that way, pure python code can be injected into the c++ inference.
This functor visualizes the labeling as an image during inference.
Args :
shape : shape of the image
numLabels : number of labels
"""
def __init__(self,shape,numLabels):
self.shape=shape
self.numLabels=numLabels
matplotlib.interactive(True)
def begin(self,inference):
"""
this function is called from c++ when inference is started
Args :
inference : python wrapped c++ solver which is passed from c++
"""
print "begin"
def end(self,inference):
"""
this function is called from c++ when inference ends
Args :
inference : python wrapped c++ solver which is passed from c++
"""
print "end"
def visit(self,inference):
"""
this function is called from c++ each time the visitor is called
Args :
inference : python wrapped c++ solver which is passed from c++
"""
arg = inference.arg()
gm = inference.gm()
print "energy ",gm.evaluate(arg)
arg=arg.reshape(self.shape)*255
plt.imshow(arg.T, cmap='gray',interpolation="nearest")
plt.draw()
def denoiseModel(
img,
norm = 2,
weight = 1.0,
truncate = None,
numLabels = 256,
neighbourhood = 4,
inpaintPixels = None,
randInpaitStartingPoint = False
):
"""
this function is used to set up a graphical model similar to
**Denoising and inpainting problems:** from `Mrf- Benchmark <http://vision.middlebury.edu/MRF/results/ >`_
Args :
img : a grayscale image in the range [0,256)
norm : used norm for unaries and 2-order functions (default : 2)
weight : weight of 2-order functions (default : 1.0)
truncate : Truncate second order function at an given value (defaut : None)
numLabels : number of labels for each variable in the graphical model,
set this to a lower number to speed up inference (default : 255)
neighbourhood : neighbourhood for the second order functions, so far only 4 is allowed (default : 4)
inpaintPixels : a tuple of x and y coordinates where no unaries are added
randInpaitStartingPoint : use a random starting point for all pixels without unaries (default : False)
"""
shape = img.shape
if(img.ndim!=2):
raise RuntimeError("image must be gray")
if neighbourhood != 4 :
raise RuntimeError("A neighbourhood other than 4 is not yet implemented")
# normalize and flatten image
iMin = numpy.min(img)
iMax = numpy.max(img)
imgNorm = ((img[:,:]-iMin)/(iMax-iMin))*float(numLabels)
imgFlat = imgNorm.reshape(-1).astype(numpy.uint64)
# Set up Grapical Model:
numVar = int(img.size)
gm = opengm.gm([numLabels]*numVar,operator='adder')
gm.reserveFunctions(numLabels,'explicit')
numberOfPairwiseFactors=shape[0]*(shape[1]-1) + shape[1]*(shape[0]-1)
gm.reserveFactors(numVar-len(inpaintPixels[0]) + numberOfPairwiseFactors )
# Set up unaries:
# - create a range of all possible labels
allPossiblePixelValues=numpy.arange(numLabels)
pixelValueRep = numpy.repeat(allPossiblePixelValues[:,numpy.newaxis],numLabels,1)
# - repeat [0,1,2,3,...,253,254,255] numVar times
labelRange = numpy.arange(numLabels,dtype=opengm.value_type)
labelRange = numpy.repeat(labelRange[numpy.newaxis,:], numLabels, 0)
unaries = numpy.abs(pixelValueRep - labelRange)**norm
# - add unaries to the graphical model
fids=gm.addFunctions(unaries.astype(opengm.value_type))
# add unary factors to graphical model
if(inpaintPixels is None):
for l in xrange(numLabels):
whereL=numpy.where(imgFlat==l)
gm.addFactors(fids[l],whereL[0].astype(opengm.index_type))
else:
# get vis of inpaint pixels
ipX = inpaintPixels[0]
ipY = inpaintPixels[1]
ipVi = ipX*shape[1] + ipY
for l in xrange(numLabels):
whereL=numpy.where(imgFlat==l)
notInInpaint=numpy.setdiff1d(whereL[0],ipVi)
gm.addFactors(fids[l],notInInpaint.astype(opengm.index_type))
# add ONE second order function
f=opengm.differenceFunction(shape=[numLabels,numLabels],norm=2,weight=weight,truncate=truncate)
fid=gm.addFunction(f)
vis2Order=opengm.secondOrderGridVis(shape[0],shape[1],True)
# add all second order factors
gm.addFactors(fid,vis2Order)
# create a starting point
startingPoint = imgFlat.copy()
if randInpaitStartingPoint :
startingPointRandom = numpy.random.randint(0,numLabels,size=numVar).astype(opengm.index_type)
ipVi = inpaintPixels[0]*shape[1] + inpaintPixels[1]
for x in ipVi:
startingPoint[x]=startingPointRandom[x]
startingPoint[startingPoint==numLabels]=numLabels-1
return gm,startingPoint.astype(opengm.index_type)
if __name__ == "__main__":
# setup
imgPath = 'houseM-input.png'
norm = 2
weight = 5.0
numLabels = 50 # use 256 for full-model (slow)
# Read image
img = numpy.array(numpy.squeeze(vigra.impex.readImage(imgPath)),dtype=opengm.value_type)#[0:100,0:40]
shape = img.shape
# get graphical model an starting point
gm,startingPoint=denoiseModel(img,norm=norm,weight=weight,inpaintPixels=numpy.where(img==0),
numLabels=numLabels,randInpaitStartingPoint=True)
inf=opengm.inference.Imc(gm,parameter=opengm.InfParam())
print "inf"
inf.setStartingPoint(inf.arg())
# set up visitor
callback=PyCallback(shape,numLabels)
visitor=inf.pythonVisitor(callback,visitNth=1)
inf.infer(visitor)
# get the result
arg=inf.arg()
arg=arg.reshape(shape)
# plot final result
matplotlib.interactive(False)
# Two subplots, the axes array is 1-d
f, axarr = plt.subplots(1,2)
axarr[0].imshow(img.T, cmap = cm.Greys_r)
axarr[0].set_title('Input Image')
axarr[1].imshow(arg.T, cmap = cm.Greys_r)
axarr[1].set_title('Solution')
plt.show()
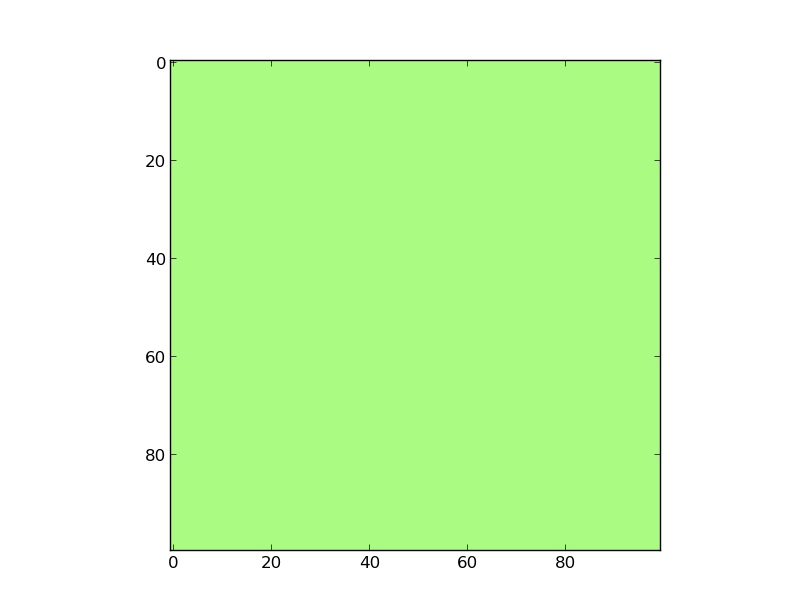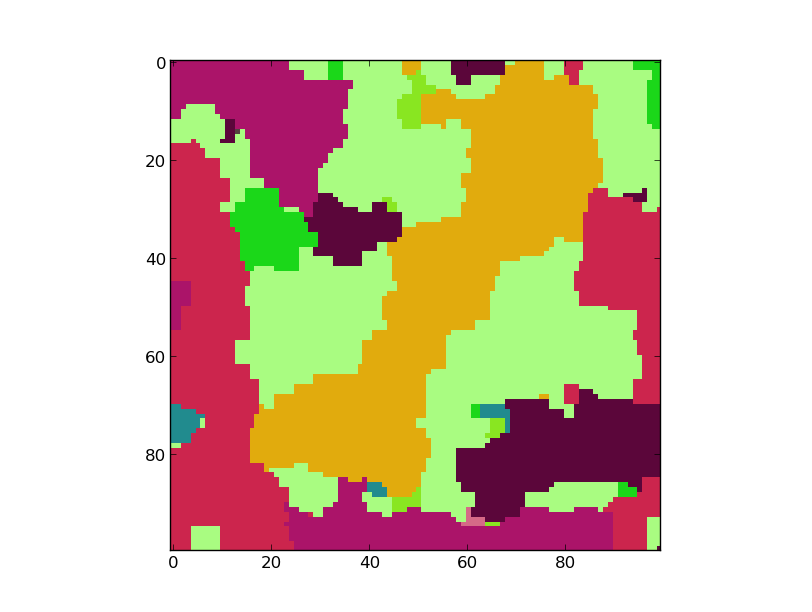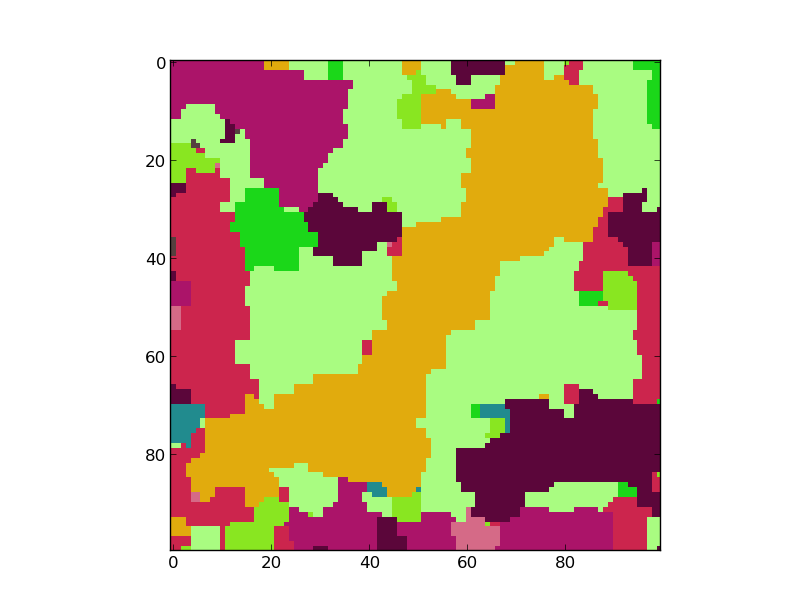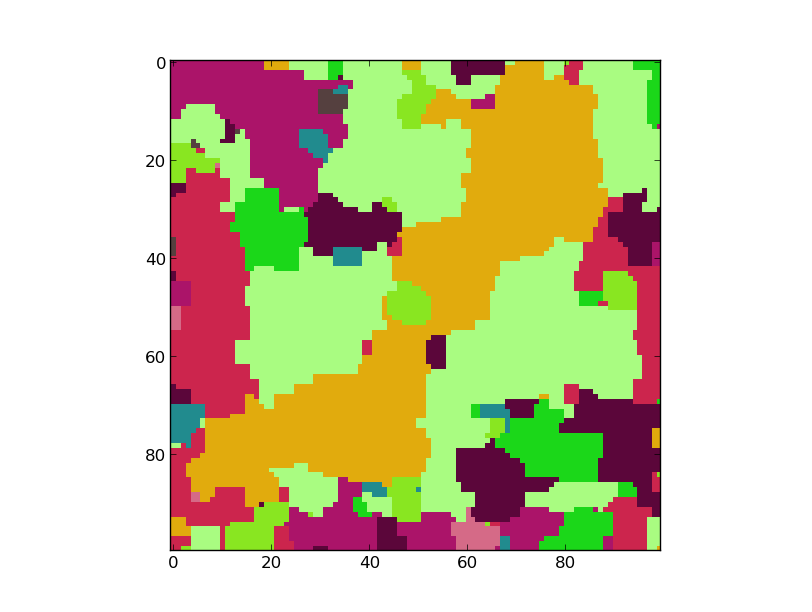
Result:¶
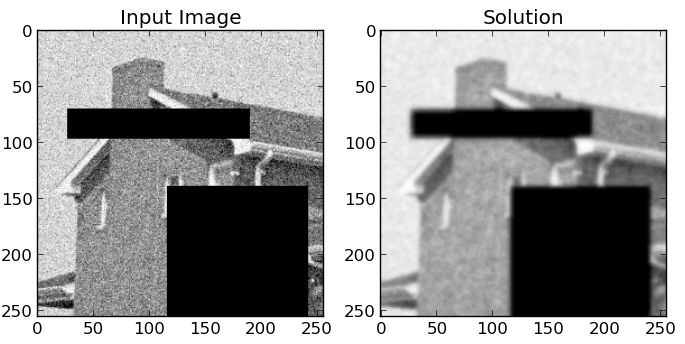
This is the result if icm is used as a solver. The model has 50 Labels, and a weight of 5.0. No truncation has been used.